
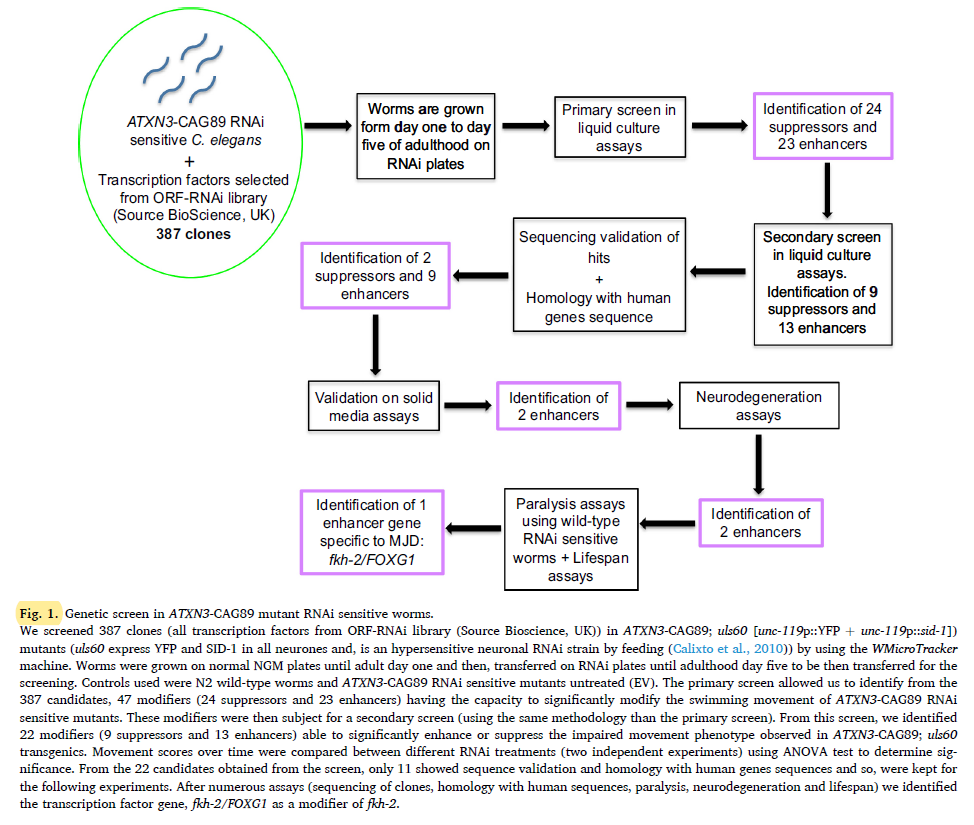
Machado-Joseph disease (MJD), also known as spinocerebellar ataxia type 3 (SCA3), is the most common form of dominantly inherited ataxia worldwide. This disease is caused by an expanded CAG repeat in the coding region of ATXN3. Due to our incomplete understanding of mechanisms and molecular pathways, there are no therapies that successfully treat core MJD patients. Therefore, the identification of new candidate targets related to this disease is needed. In this study, we performed a large-scale RNA interference (RNAi) screen of 387 transcription factor genes leading to the identification of several modifiers (suppressors and enhancers) of impaired motility phenotypes in a mutant ATXN3 transgenic C. elegans model.
Worms were exposed from adult day one to adult day five on corresponding RNAi plates and then, were transferred into a 96-well-microtiter plate. Each well contained a final volume of 100 μL of M9 buffer and ~ 50 worms adult day five obtained by synchronization. The nematodes swimming movements were tracked for ten hours using the WMicroTracker device. Two independent screening experiments were completed in ATXN3-CAG89; uIs60 worms (mutant ATXN3 transgenic strain that also contained a RNAi-sensitizing transgene) in order to identify modifiers (suppressors and/or enhancers) involved in MJD. Measurements were performed in triplicates and the average movement score was compared to the untreated ATXN3-CAG89;uIs60 mutants (EV control). If the values were higher than the control for a certain RNAi, then a secondary screen was performed to validate our observations. Candidates that modified (suppressed and/or enhanced) significantly the swimming movement of ATXN3-CAG89; uIs60 worms in the secondary screen were considered as hit candidates and so, were investigated and validated on other type of approaches.
The primary screen allowed us to identify from the 387 candidates, 47 modifiers (24 suppressors and 23 enhancers) having the capacity to significantly modify the swimming movement of ATXN3-CAG89 RNAi sensitive mutants. These modifiers were then subject for a secondary screen (using the same methodology than the primary screen). From this screen, we identified 22 modifiers (9 suppressors and 13 enhancers) able to significantly enhance or suppress the impaired movement phenotype observed in ATXN3-CAG89; uls60 transgenic. These candidates were then evaluated by sequence validation and homology with human genes sequences. From these, 11 modifiers (2 suppressors and 9 enhancers) respecting these two parameters were kept for further analysis.
Exp Neurol. 2021 Mar;337:113544. doi: 10.1016/j.expneurol.2020.113544. Epub 2020 Dec 5.
Yasmin Fardghassemi, Alex Parker.
