There is a continuous need for the development of simple animal models for the study of host-pathogen interactions. There is also a need for the identification and study of virulence mechanisms as well. Several features of C. elegans make it a powerful model organism to study host-pathogen interaction at genetic and molecular level. C. elegans share conserved pathogen-induced virulence (e.g. Staphylococcus V8 protease, Pseudomonas exotoxin A) and host innate immune mechanisms (e.g. antimicrobial peptides (AMPs) and MAPK pathways) with mammals, arthropods and plants (Kumar et al 2019).
Caenorhabditis elegans can be killed by diverse range of human pathogens, such as Gram-positive (Enterococcus faecalis, Staphylococcus aureus, and Streptococcus pneumoniae) and Gram-negative bacteria (Salmonella enterica, Pseudomonas aeruginosa, Burkholderia cepacia, and Serratia marcescens) as well as different fungi (Candida albicans and C. neoformans) (Kumar et al 2019).
Generally, the bacterial pathogens infect nematodes by direct colonization within the intestine or indirectly by secretion of toxins. Using WMicrotracker, it is possible to easily study bacterial infection in C.elegans (and other small organisms) in order to discover novel antibiotics, natural killers, or genes that improve immune resistance. Below we present an example of application:
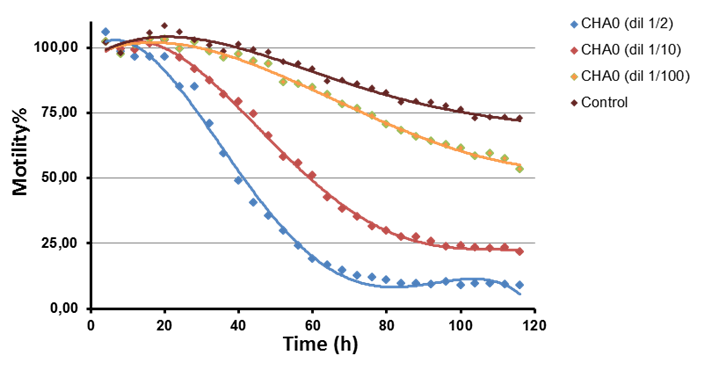
In this experiment we can observe long-term kinetic and dose response effect using dilutions of bacterial supernatant of Pseudomonas CHA0. Paralytic killing is reported to depend on bacterial hydrogen cyanide production
PROTOCOL:
- Obtain synchronized populations of adult GLP-4 worms grown in NGM + OP50 at 25°C.
- Remove worms from plates using M9 buffer and transfer them in a sterile 15 ml tube.
- Let the worms settle. Decant the supernatant taking care not to disturb the pellet.
- Perform a wash with 5 ml of M9 buffer. Briefly shake or invert the tube.
- Add 3 ml of nutrient medium
- Count the number of worms in a volume of 10 µl and adjust the volume to obtain a concentration of [5 worms / 10 µl].
- Transfer per well 90 μl of the worm solution to a 96 well microplate using multichannel pipette.
- Add 10 µl of a bacterial culture supernatant to test and gently shake microplate by hand.
- Record the activity of the plate with worms using WMicrotracker.
- Generate the data report using WMicrotracker One software and plot using MSExcel.
Cell Mol Life Sci. 2020 Apr;77(7):1229-1249. doi: 10.1007/s00018-019-03319-7.
Kumar A, Baruah A, Tomioka M, Iino Y, Kalita MC, Khan M.
